Influenza Co-Infection in COVID-19 Patients
Influenza Co-Infection in COVID-19 Patients
Caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus, coronavirus disease 2019 (COVID-19) is a major public health burden which can be devastating 1. Influenza on the other hand remains as one of the world’s greatest health concerns, with an estimated 1 billion cases every year. Of these cases, 3 – 5 million are severe, resulting in approximately 290,000 – 650,000 deaths per year 2. Whilst influenza is commonly discussed with regards to past or future pandemics, seasonal influenza is responsible for the substantial burden of lower respiratory tract infections (LRTIs) and other respiratory conditions 3.
The measures put in place to reduce the transmission of COVID-19 has also been effective in reducing the transmission of other respiratory viruses, such as influenza. With the measures decreased as we continue with a new normal, it is expected that COVID-19 will circulate with other respiratory viruses, increasing the likelihood of co-infections. Co-infection with influenza is associated with an increased risk of receiving invasive mechanical ventilation compared with COVID-19 mono-infection. SARS-CoV-2 co-infection with influenza is significantly associated with an increased risk of adverse outcomes and death 4.
Host and viral factors determine the extent and severity of virus-induced lung damage. The host’s response to the viral infection is essential for viral clearance, however, the host’s response could be damaging and could contribute towards disease severity. Likewise, the mechanisms of tissue repair is essential for recovery from the infection, across the spectrum of disease severity, but, dysregulated repair responses could lead to chronic lung dysfunction. Consequently, it is vital to understand the mechanisms of immunopathology and tissue repair following LRT viral infection as it could aid in effective treatment plan implementation 5.
Pathiophysiology
Influenza
Influenza belongs to the Orthomyxoviridae family of negative-sense RNA viruses. Four types of influenza have been discovered; A (IAV), B (IBV), C (ICV) and D (IDV) 6.
Figure 1 summarises the natural history, pathogenesis and replication cycle of influenza viruses. IAV, IBV and ICV infect humans, however, the high mutation rates enables the invasion of immunity. IAV from different host species is capable of ‘reassorting’ their segmented genomes, creating pandemic strains that are antigenically novel but well adapted to humans 7.
In humans, the influenza viruses infect the respiratory epithelium. Sialic acid is bound by the haemagglutinin (HA) proteins of IAV and IBV, or the haemagglutinin-esterase-fusion (HEF) proteins of ICV, causing endocytosis. The viral genome of RNA viruses replicate in the nucleus. New viruses gather at the cell surface and are released by the receptor-cleaving neuraminidase (NA) proteins of IAV and IBV or the ICV HEF proteins 7. Transmission of the virus occurs when a susceptible individual comes into contact with fomites from an infected individual or with aerosols 8.
Figure 1: The natural history, pathogenesis and replication cycle of influenza viruses 7
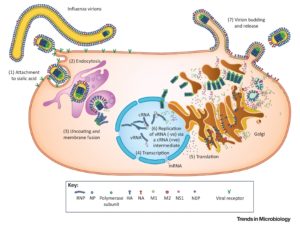
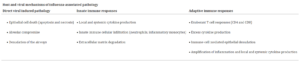
The pathophysiology of influenza is primarily lung inflammation caused by immune responses employed to handle the spread of infection, combined with lung compromise caused by the direct viral infection of the respiratory epithelium (see table 1) 8.
Table 1: Host and viral mechanisms of influenza-associated pathology 8
SARS-COV-2
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to the Coronaviridae family of coronaviruses, which are common pathogens in animals and humans. Four human coronaviruses have been discovered; HCoV-NL63, HCoV-229E, HCoV-OC43 and HCoV-HKU1. These coronaviruses affect the upper respiratory tract, producing common-cold like symptoms. Three zoonotic coronaviruses have been discovered: SARS-CoV, SARS-CoV-2 and MERS-CoV (Middle East Respiratory Syndrome). These zoonotic coronaviruses have affected humans upon leaking over from animal reservoirs 9.
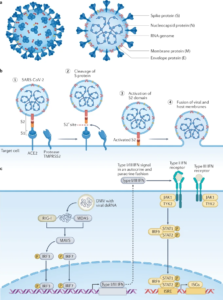
SARS-CoV-2 belongs to the genus Sarbecovirus and shares a sequence similarity of 79% with SARS-CoV. SARS-CoV-2 encodes a set of accessory proteins, structural proteins (envelope protein, membrane protein, nucleocapsid protein and spike glycoprotein) and non-structural proteins (most compose the viral replication and transcription complex). A lipid bilayer from the host and the structural proteins form and enveloped virion which delivers viral genomic RNA into the cell (see figure 2a). The accessory proteins are dispensed for replication, however, they can often have immunoevasive activities. The spike glycoprotein, which forms trimers on the surface of virions is the main determinant of the coronavirus tropism. The S protein attaches to the receptor angiotensin-converting enzyme 2 (ACE2) on the host cell using the S1 subunit, enabling transmembrane protease serine 2 (TMPRSS2) to cleave to the S protein, activating the S2 subunit for fusion with the host lipid bilayers. This results in the reposition of the viral positive-sense, single-stranded RNA genome into the host cell (figure 2b). The nasopharynx or trachea cells are the first cells targeted by SARS-CoV-2 during natural infection in humans. Upon entry, the positive-sense SARS-CoV-2 genome directly instigates the production of viral proteins, including the replicase proteins that form replication factories which contain double membrane vesicles (where transcription occurs) from the endoplasmic reticulum membranes. This shields the double-stranded RNA (dsRNA) transcription intermediates from detection by cytoplasmic pattern recognition receptors (PRRs) 9.
Figure 2: Molecular and Cellular Pathogenesis of SARS-CoV-2 9
Co-Infection Testing
The co-circulation of the influenza virus merging with the COVID-19 pandemic raises a potentially severe threat to public health. SARS-CoV-2 co-infection with other pathogens may worsen the severity and clinical outcome of COVID-19 and increase fatality 10.
Research undertaken by the International Severe Acute Respiratory and Emerging Infection Consortium examined the clinical outcomes of COVID-19 co-infection with influenza, respiratory syncytial virus (RSV) and adenoviruses in 212, 366 adults with SARS-CoV-2 who were admitted to hospital in the UK between 6th February 2020 and 8th December 2021. 6,965 patients were tested for respiratory viral co-infection of which 583 (8.3%) patients tested positive: 227 patients had influenza, 220 patients had RSV and 136 patients had adenovirus. SARS-CoV-2 co-infection with adenoviruses or influenza were significantly associated with increased odds of death and co-infection with influenza was associated with increased odds of receiving invasive mechanical ventilation compared with SARS-CoV-2 mono-infection (see table 2) 4.
Table 2: Multivariable Model of the Effect of Co-Infection Compared with SARS-CoV-2 Mono-infection 4
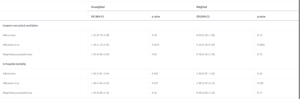
As the public health restrictions are lifted, respiratory virus co-infections are highly likely to occur during upcoming winters. The results of the above study highlight the importance of testing for influenza among hospital in-patients with COVID-19 to identify at risk patients 4.
A systemic review and meta-analysis was undertaken, retrieving 6,053 publications of patients who presented with COVID-19 and were screened for a secondary respiratory illnesses. 59 studies with a total of 16,643 SARS-CoV-2 positive patients were included. The global pooled prevalence was 5.01% based on the random-effects model, with influenza (1.54%) and enteroviruses (1.32%) being the most prevalent pathogens. The subgroup analysis found that co-infection was significantly higher in paediatrics (9.39%) compared to adults (3.51%). These results highlight the need for multiplex viral testing in in-patients with compatible symptoms 11.
Biorex Diagnostics COVID/Influenza Product Portfolio
| Description | Cat No. | Size | Methodology |
| COVID-19 IgG/IgM Rapid Test Cassette | COVID010 | 10T | Rapid |
| COVID-19 IgG/IgM Rapid Test Cassette | COVID020 | 20T | Rapid |
| COVID-19 Ag Nasal/Nasopharyngeal Swab Cassette | COVNS020 | 20T | Rapid |
| Influenza & COVID-19 Ag Combo Rapid Test (Nasopharyngeal) | COVIC020 | 20T | Rapid |
| Influenza & COVID-19 Ag Combo Rapid Test (Nasal) | COVICN20 | 20T | Rapid |
| COVID-19 Total Antibody | BXE0882A
|
96T | ELISA |
| COVID-19 IgM Antibody | BXE0883A | 96T | ELISA |
| SARS-CoV-2 Neutralising Antibody (IgG) Antibody | BXE0885A
|
96T | ELISA |
References
- R.J. Pathogenesis of COVID-19 from a cell biology perspective. European Respiratory Journal 2020; 55(4): . https://erj.ersjournals.com/content/55/4/2000607 (accessed 27 July 2022).
- C. WHO launches new global influenza strategy. https://www.who.int/news-room/detail/11-03-2019-who-launches-new-global-influenza-strategy (accessed 27 July 22).
- Troeger CE, Blacker BF, Khalil IA, Zimsen SRM, Albertson SB, Abate D. Mortality, morbidity, and hospitalisations due to influenza lower respiratory tract infections, 2017: an analysis for the Global Burden of Disease Study 2017. The Lancet Respiratory Medicine 2019; 7(1): . https://www.thelancet.com/journals/lanres/article/PIIS2213-2600(18)30496-X/fulltext#%20 (accessed 27 July 2022).
- Swets MC, Russell CD, Harrison EM, Docherty AB, Lone N, Girvan M. SARS-CoV-2 co-infection with influenza viruses, respiratory syncytial virus, or adenoviruses. The Lancet 2022; 399(10334): . https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(22)00383-X/fulltext#:~:text=Co%2Dinfection%20with%20influenaza%20viruses,with%20increased%20odds%20of%20death (accessed 27 July 2022).
- Flerlage T, Boyd DF, Meliopoulos V, Thomas PG, Schultz-Cherry S. Influenza virus and SARS-CoV-2: pathogenesis and host responses in the respiratory tract. Nature Reviews Microbiology 2021; 19(): . https://www.nature.com/articles/s41579-021-00542-7 (accessed 22 August 2022).
- Gounder AP, Boon ACM. Influenza Pathogenesis: The Effect of Host Factors on Severity of Disease. The Journal of Immunology 2019; 202(2): . https://www.jimmunol.org/content/202/2/341 (accessed 22 August 2022).
- Hutchinson EC. Influenza Virus. Trends in Microbiology 2018; 26(9): . https://www.cell.com/trends/microbiology/fulltext/S0966-842X(18)30131-8 (accessed 23 August 2022).
- Kalil AC, Thomas PG. Influenza virus-related critical illness: pathophysiology and epidemiology. Critical Care 2019; 23(7): . https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6642581/ (accessed 23 August 2022).
- Lamers MM, Haagmans BL. SARS-CoV-2 pathogenesis. Nature Reviews Microbiology 2022; 20(): . https://www.nature.com/articles/s41579-022-00713-0 (accessed 23 August 2022).
- Kin EH. Hguyen TQ, Casel MAB, Rollon R, Kim SM, et al. Coinfection with SARS-CoV-2 and Influenza A Virus Increases Disease Severity and Impairs Neutralizing Antibody and CD4+ T Cell Responses. Journal of Virology 2022; 96(6): . https://journals.asm.org/doi/10.1128/jvi.01873-21 (accessed 21 September 2022).
- Krumbein H, Kümmel LS, Fragkou PC, Thölken C, Hünerbein BL, et al. Respiratory viral co-infections in patients with COVID-19 and associated outcomes: A systemic review and meta-analysis. Reviews in Medical Virology 2022; e2365(): . https://onlinelibrary.wiley.com/doi/epdf/10.1002/rmv.2365 (accessed 21 September 2022).